DAAO
D-Amino acid oxidase
D-Amino acid oxidase (DAAO, EC 1.4.3.3) is a flavin adenine dinucleotide (FAD)-containing enzyme belonging to the dehydrogenase/oxidase class of flavoproteins. It catalyzes the dehydrogenation of the D-isomer of amino acids to the corresponding imino acids, coupled with the reduction of FAD to FADH2. The imino acid then hydrolyzes spontaneously to give α-keto acid and ammonia; the reduced flavin cofactor reoxidizes on molecular oxygen, producing hydrogen peroxide (Fig. 1) [Pollegioni et al., 2008].
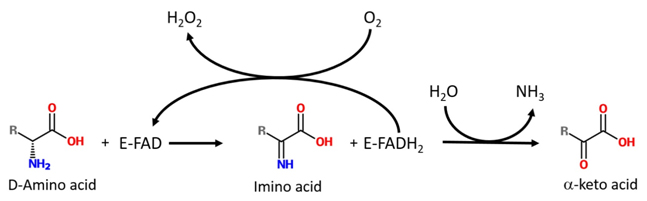
DAAO shows an absolute stereospecificity (it does not oxidize L-AAs) and a broad substrate specificity. It oxidizes aliphatic, aromatic, and polar D-AAs, whereas D-aspartic acid and D-glutamic acid are not substrates for DAAO [Pollegioni et al., 2008]. Instead, these two latter compounds are substrates of D-aspartate oxidase (DASPO).
DAAO was discovered in 1935 by Sir Hans Krebs and since then it has been the subject of a great mass of structural, functional, and kinetic investigations (reviewed in [Curti et al., 1992; Pilone, 2000]) that make DAAO a model enzyme for the dehydrogenase/oxidase class of flavoproteins. A fundamental advance in the comprehension of the structure-function relationships in DAAO was made with the resolution of the 3D-structure of the enzyme from pig kidney (pkDAAO) [Mattevi et al., 1996; Mizutani et al., 1996], from the yeast Rhodotorula gracilis (RgDAAO) [Umhau et al., 2000], and from human (hDAAO) [Kawazoe et al., 2006]. A second fundamental factor that allowed apparent advances in DAAO studies was the availability of DAAO genes/cDNAs from different organisms. Owing to both of these aspects, protein engineering techniques such as site-directed mutagenesis and directed evolution could be applied [Pollegioni et al., 2007b; Pollegioni and Molla, 2011].
Distribution and physiological role(s)
DAAO is a peroxisomal enzyme present in all eukaryotic taxa, with the exception of plants; it is involved in a variety of different physiological functions (for an exhaustive review, see [Pollegioni et al., 2007a]). In yeasts, for example, oxidation of D-AAs makes possible their use as carbon, nitrogen, or energy source. In animals (both invertebrates and vertebrates), the role of DAAO is related to species-specific functions. In mammals, DAAO is present in kidney, where its role is related to eliminating D-AAs deriving from spontaneous racemization, diet, or bacterial sources. However, a main role of mammalian (and especially human) DAAO is related to its presence in selected brain areas; here, it catabolizes D-serine, a neuromodulator that acts as a coagonist of the N-methyl-D-aspartate receptors (NMDAr). Indeed, NMDAr dysfunction has been correlated to different neurodegenerative or psychiatric diseases, such as familial amyotrophic lateral sclerosis, Alzheimer’s disease, and schizophrenia [Pollegioni and Sacchi, 2010; Sacchi et al., 2012]. Thus, by modulating D-serine levels, human DAAO plays a key role in regulating the activation state of NMDAr. As a result, the role of DAAO in human brain studies focused on the discovery of new drugs acting as DAAO inhibitors/modulators for the treatment of the aforementioned diseases.
Biochemical and structural properties
Yeast and mammalian DAAOs show about 30% sequence identity, whereas pkDAAO shares an 85% sequence identity with hDAAO. Despite the fact that DAAO from different sources share the same catalytic mechanism, they show important differences in many biochemical properties, too, including catalytic efficiency, substrate specificity, oligomeric state, stability, kinetic mechanism, and FAD binding. All DAAOs bind the FAD cofactor in a noncovalent mode. The binding is tighter in yeast enzymes (Kd = 2×10-8 M for RgDAAO) than in mammalian enzymes (Kd = 2.2×10-7 M for pkDAAO and 8×10-6 M for hDAAO) [Pollegioni et al., 2007a; Molla et al., 2006].
DAAOs also differ in oligomeric state. RgDAAO and hDAAO are stable homodimers, whereas porcine DAAO exists in solution as a mixture of monomers, dimers, or higher oligomers, depending on the concentration and the presence of ligands [Curti et al., 1992; Pollegioni et al., 2007a]. Interestingly, only hDAAO is a homodimer even when the flavin cofactor is removed [Molla et al., 2006]: apoenzymes obtained from RgDAAO and pkDAAO are monomeric. The 3D structures of pkDAAO and hDAAO are very similar, whereas the mode of dimerization differs between mammalian and yeast enzymes. Both pkDAAO and hDAAO show a “head-to-head” dimerization mode, whereas RgDAAO shows a “head-to-tail” mode (Fig. 2).
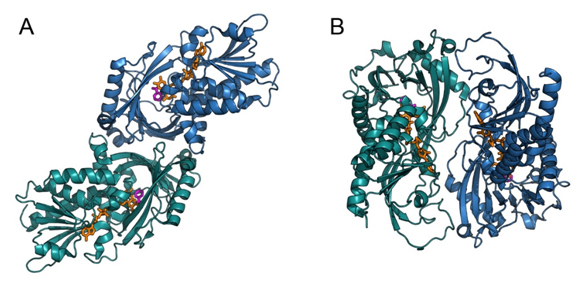
In all DAAOs, each monomer is divided into two domains [Mattevi et al., 1996; Mizutani et al., 1996; Umhau et al., 2000; Kawazoe et al., 2006]: the FAD-binding domain containing the typical βαβ dinucleotide-binding motif known as the Rossmann fold and the interface domain forming the contact surface with the second monomer. The FAD cofactor is buried inside the protein in an elongated conformation. The peculiar dimerization mode observed in RgDAAO is due to a positively charged long loop (not conserved in other DAAOs) that interacts with negatively charged residues belonging to two α-helices of the other monomer [Umhau et al., 2000; Pollegioni et al., 2002].
The different biochemical properties of DAAOs from diverse sources reflect their physiological role [Pollegioni et al., 2007a]: an efficient catabolic enzyme in yeasts and an enzyme able to finely tune the levels of the neuromodulator D-Ser in mammalian brain.
Biotechnological applications of DAAO
Microbial DAAOs possess properties that render them suitable for biotechnological applications. They are stable enzymes and show broad substrate specificity, high turnover number, and a tight binding with the FAD cofactor. Moreover, they can be produced at high levels, both as native and recombinant proteins. This made possible the production of enzyme variants with new and evolved properties by using protein engineering techniques on the basis of knowledge of the structure-function relationships. The main biotechnological applications of DAAO have been reviewed in detail in [Pollegioni et al., 2007b, 2008, 2011] and comprise: a) production of 7-amino cephalosporanic acid (7-ACA) from cephalosporin C (CephC); b) resolution of racemic solutions of amino acids; c) detection and quantification of D-AAs in biological samples; and d) selective marker in plants.
Production of antibiotics
7-ACA is the starting point for producing a number of semi-synthetic cephalosporins. 7-ACA is industrially produced in a well-established, two-step, enzymatic process starting from CephC: the first step is the conversion of CephC into glutaryl-7-amino cephalosporanic acid (Gl-7-ACA) catalyzed by DAAO; then, the enzyme glutaryl acylase converts Gl-7-ACA into 7-ACA [Pilone, 2000; Pollegioni et al., 2008]. This process is currently the most important industrial application of DAAO. Over the years, some improvements in the process have been made by protein engineering studies. These studies include: a) site-directed mutagenesis of Trigonopsis variabilis DAAO (TvDAAO) to obtain a 6-fold increased maximal activity [Wong et al. 2010]; b) production of RgDAAO and TvDAAO variants fused with hemoglobin [Khang et al., 2003; Ma et al., 2009] or more active at low O2 concentration [Rosini et al., 2009; Rosini et al., 2010]; c) production of a chimeric enzyme made of Pseudomonas glutaryl acylase and TvDAAO [Luo et al., 2004]; and d) coexpression of both DAAO and glutaryl acylase in E. coli cells [Zheng et al., 2007 ]. In c) and d) a so-called “one-pot” (i.e., a single reactor) conversion of CephC into 7-ACA was obtained.
Resolution of racemic mixtures of amino acids
Availability of enantiomerically pure α-amino acids or unnatural amino acids is of increasing interest for the fine chemical and pharmaceutical industries, especially for drug discovery. This aim was supported by the rational design of RgDAAO together with the use of multi-enzymatic systems. The use of the M213G RgDAAO variant allowed the complete resolution of racemic unnatural amino acids tested in a short time (up to 10-times faster) and using low amounts of enzyme (down to 60-fold less) compared to the wild-type counterpart [Caligiuri et al., 2006a]. The complete recovery of L-enantiomers was obtained by combining the M213G RgDAAO variant with L-aspartate amino transferase and catalase [Caligiuri et al., 2006b]. Similarly, in a four-enzyme system including DAAO from Arthrobacter protophormiae, D-methionine was fully converted to the L-enantiomer [Findrik and Vasić-Racki, 2007].
DAAO-based biosensors
Detection of D-AAs in biological samples is an important issue: their presence in food (especially in dairy products) indicates bacterial contamination (D-AAs are components of the bacterial cell wall) and D-serine is a neuromodulator acting on NMDA receptors in the mammalian brain (see above).
Biosensors for different D-AAs based on commercial pkDAAO appeared between 2001 and 2003. DAAO was employed in combination with horseradish peroxidase to determine several common D-AAs [Domínguez et al., 2001] and D-pipecolic acid (a marker for peroxisomal disorders) [Stefan et al., 2003a], or in combination with pyruvate oxidase to determine D-alanine [Inaba et al., 2003]. DAAO-based biosensors have been also employed to identify clinically relevant molecules such as D-methotrexate [Stefan et al., 2003b] and D-thyroxine [van Staden et al., 2010].
Significantly improved biosensors in terms of sensitivity and substrate specificity have been obtained by substituting commercial pkDAAO with RgDAAO. This allowed D-serine levels to be determined in vivo [Pernot et al., 2008; Polcari et al., 2014] and, using an evolved RgDAAO obtained by combining rational design and directed-evolution approaches, the total concentration of all D-AAs present in a biological sample to be measured [Rosini et al., 2008]. Very recently, a bienzymatic biosensor based on commercial pkDAAO and horse radish peroxidase coimmobilized on carbon nanotubes and gold nanoparticles was set up. With this innovative device total D-AA content can be detected in complex biological matrices, such as bacterial samples [Moreno-Guzmán et al., 2017].
Use of DAAO in agriculture
Due to the absence of DAAO activity in plants, it can be used as an innovative system both for positive and negative selection of transgenic plants. This strategy was applied for the first time to an economically relevant species (apple) in 2009 [Hättasch et al., 2009]. A second application of DAAO in agriculture is the production of hybrid F1 seeds. For this purpose, the male reproductive organs must be selectively destroyed to avoid undesired self-pollination. To this aim, an evolved variant of Rhodosporidium toruloides DAAO (RtDAAO) has been employed in tobacco plants. This system is based on the conversion of the inactive D-enantiomer of the herbicide glufosinate into its toxic L-enantiomer by the evolved RtDAAO (selectively expressed in anthers) in conjunction with the endogenous enzyme L-glutamate aminotransferase [Hawkes et al., 2011]. Evolved RtDAAO is much more active on D-glufosinate with respect to the wild-type enzyme and the combined action of the two enzymes produces transgenic lines, exhibiting complete male sterility that persists for more than two weeks after foliar treatment with D-glufosinate [Hawkes et al., 2011].
Links
Brenda (http://www.brenda-enzymes.org/enzyme.php?ecno=1.4.3.1)
Wikipedia (https://en.wikipedia.org/wiki/D-amino_acid_oxidase)
ExPASy (http://enzyme.expasy.org/EC/1.4.3.3)
UniProt (human) (http://www.uniprot.org/uniprot/P14920)
UniProt (pig) (http://www.uniprot.org/uniprot/P00371)
UniProt (R. gracilis) (http://www.uniprot.org/uniprot/P80324)
References
Caligiuri A., D’Arrigo P., Rosini E., Tessaro D., Molla G., Servi S., Pollegioni L. (2006a) Enzymatic conversion of unnatural amino acids by yeast D-amino acid oxidase. Adv. Synth. Catal. 348, 2183-2190.
Caligiuri A., D’Arrigo P., Gefflaut T., Molla G., Pollegioni L., Rosini E., Rossi C., Servi S. (2006b) Multistep enzyme catalysed deracemisation of 2-naphthylalanine. Biocatal. Biotransform. 24, 409-413.
Curti B., Ronchi S., Pilone M.S. (1992) D- and L-amino acid oxidase. In: Chemistry and Biochemistry of Flavoproteins, vol. 3, pp. 69-94, Müller F. (Ed.), CRC Press, Boca Raton (FL).
Domínguez R., Serra A., Reviejo A.J., Pingarrón J.M. (2001) Chiral analysis of amino acids using electrochemical composite bienzyme biosensors. Anal. Biochem. 298, 275-282.
Findrik Z., Vasić-Racki D. (2007) Biotransformation of D-methionine into L-methionine in the cascade of four enzymes. Biotechnol. Bioeng. 98(5), 956-967.
Hawkes T., Pline-Srnic W., Dale R., Friend E., Hollinshead T., Howe P., Thompson P., Viner R., Greenland A. (2011) D-glufosinate as a male sterility agent for hybrid seed production. Plant Biotechol. J. 9, 301-314.
Hättasch C., Hanke M.-V., Flachowsky H. (2009) Preliminary results to establish the DAAO system as an alternative selection strategy on apple. Acta Horticulturae 814, 267-272.
Inaba Y., Mizukami K., Hamada-Sato N., Kobayashi T., Imada C., Watanabe E. (2003) Development of a D-alanine sensor for the monitoring of a fermentation using the improved selectivity by the combination of D-amino acid oxidase and pyruvate oxidase. Biosens. Bioelectron. 19, 423-431.
Kawazoe T., Tsuge H., Pilone M.S., Fukui K. (2006) Crystal structure of human D-amino acid oxidase: context-dependent variability of the backbone conformation of the VAAGL hydrophobic stretch located at the si-face of the flavin ring. Protein Sci. 15(12), 2708-2717.
Khang Y.-H., Kim I.-W., Hah Y.-R., Hwangbo J.-H., Kang K.-K. (2003) Fusion protein of Vitreoscilla hemoglobin with D-amino acid oxidase enhances activity and stability of biocatalyst in the bioconversion process of cephalosporin C. Biotechnol. Bioeng. 82, 480-488.
Luo H., Li Q., Yu H., Shen Z. (2004) Construction and application of fusion proteins of D-amino acid oxidase and glutaryl-7-aminocephalosporanic acid acylase for direct bioconversion of cephalosporin C to 7-aminocephalosporanic acid. Biotechnol. Lett. 26, 939-945.
Ma X.-F., Yu H.-M., Wen C., Luo H., Li Q., Shen Z.-Y. (2009) Triple fusion of D-amino acid oxidase Trigonopsis variabilis with polyhistidine and Vitreoscilla hemoglobin. World J. Microb. Biotechnol. 25,1353-1361.
Mattevi A., Vanoni M.A., Todone F., Rizzi M., Teplyakov A., Coda A., Bolognesi M., Curti B. (1996) Crystal structure of D-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b2. Proc. Natl. Acad. Sci. USA 93(15), 7496-7501.
Mizutani H., Miyahara I, Hirotsu K., Nishina Y., Shiga K., Setoyama C., Miura R. (1996) Three-dimensional structure of porcine kidney D-amino acid oxidase at 3.0 A resolution. J. Biochem. (Tokyo) 120(1), 14-17.
Molla G., Sacchi S., Bernasconi M., Pilone M.S., Fukui K., Pollegioni L. (2006) Characterization of human D-amino acid oxidase. FEBS Lett. 580(9), 2358-2364.
Moreno-Guzmán M., García-Carmona L., Molinero-Fernández Á., Cava F., López Gil M.Á., Escarpa A. (2017) Bi-enzymatic biosensor for on-site, fast and reliable electrochemical detection of relevant D-amino acids in bacterial samples. Sens. Actuators B. Chem. 242, 95-101.
Pernot P., Mothet J.-P., Schuvailo O., Soldatkin A., Pollegioni L., Pilone M., Adeline M.T., Cespuglio R., Marinesco S. (2008) Characterization of a yeast D-amino acid oxidase microbiosensor for D-serine detection in the central nervous system. Anal. Chem. 80, 1589-1597.
Pilone M.S. (2000) D-Amino acid oxidase: new findings. Cell. Mol. Life Sci. 57, 1732-1747.
Polcari D., Kwan A., Van Horn M.R., Danis L., Pollegioni L., Ruthazer E.S., Mauzeroll J. Disk-shaped amperometric enzymatic biosensor for in vivo detection of D-serine. Anal. Chem. 86, 3501-3507.
Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G. Physiological functions of D-amino acid oxidases: from yeast to humans. Cell. Mol. Life Sci. 2007;64(11):1373-94.
Pollegioni L., Sacchi S., Caldinelli L., Boselli A., Pilone M.S., Piubelli L., Molla G. (2007b) Engineering the properties of D-amino acid oxidases by a rational and a directed evolution approach. Curr. Protein Pept. Sci. 8(6), 600-618.
Pollegioni L., Molla G., Sacchi S., Rosini E., Verga R., Pilone M.S. (2008) Properties and applications of microbial D-amino acid oxidases: current state and perspectives. Appl. Microbiol. Biotechnol. 78(1), 1-16.
Pollegioni L., Sacchi S. (2010) Metabolism of the neuromodulator D-serine. Cell. Mol. Life Sci. 67, 2387-2404.
Pollegioni L., Molla G. (2011) New biotech applications from evolved D-amino acid oxidases. Trends Biotechnol. 29(6), 276-283.
Rosini E., Molla G., Rossetti C., Pilone M.S., Pollegioni L., Sacchi S. (2008) A biosensor for all D-amino acids using evolved D-amino acid oxidase. J. Biotechnol.135, 377-384.
Rosini E., Pollegioni L., Ghisla S., Orrù R., Molla G. (2009) Optimization of D-amino acid oxidase for low substrate concentrations – towards a cancer enzyme therapy. FEBS J. 276(17), 4921-4932.
Rosini E., Molla G., Ghisla S., Pollegioni L. (2010) On the reaction of D-amino acid oxidase with dioxygen: O2 diffusion pathways and enhancement of reactivity. FEBS J. 278(3), 482-492.
Sacchi S., Caldinelli L., Cappelletti P., Pollegioni L., Molla G. (2012) Structure–function relationships in human D-amino acid oxidase. Amino Acids 43(5), 1833-1850.
Stefan R.-I., Nejema R’.M., van Staden J.F., Aboul-Enein H.Y. (2003a) Biosensors for the enantioselective analysis of pipecolic acid. Sens. Actuators B. Chem.94, 271-275.
Stefan R.-I., Bokretsion R.G., van Staden J.F., Aboul-Enein H.Y. (2003b) Simultaneous determination of L- and D-methotrexate using a sequential injection analysis/amperometric biosensors system. Biosens. Bioelectron. 19, 261-267.
van Staden R.-I., van Staden J.F., Aboul-Enein H.Y., Balcu I. (2010) Simultaneous determination of L- and D-T4 using a sequential injection analysis/sensors system. Comb. Chem. High Throughput Screen. 13(6), 497-501.
Umhau S, Pollegioni L, Molla G, Diederichs K, Welte W, Pilone MS, Ghisla S. The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation. Proc Natl Acad Sci U S A. 2000;97(23):12463-8.
Wong K.-S., Fong W.-P., Tsang P.W.-K. (2010) A single Phe54Tyr substitution improves the catalytic activity and thermostability of Trigonopsis variabilis D-amino acid oxidase. New Biotechnol. 27, 78-84.
Zheng H., Zhu T., Chen J., Zhao Y., Jiang W., Zhao G., Yang S., Yang Y. (2007) Construction of recombinant Escherichia coli D11/pMSTO and its use in enzymatic preparation of 7-aminocephalosporanic acid in one-pot. J. Biotechnol. 129(3), 400-405.
Author
Luciano Piubelli, Università degli studi dell’Insubria
